Phylogenetics, in biology, is concerned with the evolutionary relationships among biological organisms and it asserts that each species today is evolved from an ancestral species. A phylogenetic tree (Fig. 1.) is an illustration of the evolutionary history of various biological species through studying the similarities and differences in organisms (Wiley & Lieberman, 2011). Each line represents a lineage and the splitting of lineage as denoted by the node labeled “speciation event” results in two different species – sharks and osteichthyans. These two species shared a common ancestor denoted by the edge labeled “common ancestor” before the splitting of lineage happened.

Fig. 1. A biological phylogenetic tree
(Wiley & Lieberman, 2011: 5)
In linguistics too, it is assumed that languages today have evolved from ancestral languages that used to exist. There are many fundamental features that biological and linguistic evolution share (Fig. 2.).

Fig. 2. Features shared between biological and linguistic evolution
(Atkinson & Gray, 2005: 514)
The discrete characters in biological evolution, which are DNA sequences, like lexical characters, grammatical and phonological structures in languages, can be passed on from one generation to the next. Homologies in biological species, like cognates among languages indicate shared ancestry and inheritance from a unique ancestor. Cognates are words that show similar form and meaning, and can be shown to be inherited from a common ancestor through regular sound correspondences. Genes undergo changes through mutation and random drift, and so do linguistic characters. Lexical mutations can come in the form of innovation, such as the word boy, which appeared some time after English split from other west Germanic languages; as well as in the form of insertions, which happened to ‘books’ in Old Swedish which transformed from *bökr to böker, according to Campbell, 2004 (cited in Atkinson & Gray, 2005: 513). Linguistic diversification happens when lineages split forming two new languages and using this knowledge, we can reconstruct the historical relationships among languages by comparing linguistic features of languages.
Linguistic phylogenetic trees, such as in Fig. 3., represent the evolutionary history of a group of related languages (Nichols & Warnow, 2008). The common ancestor of all the daughter languages is represented by the root; the edges represents the intermediate ancestral languages; the nodes represent the splitting of language families into subfamilies; and the leaves represent the daughter languages.

Fig. 3. A phylogenetic tree
(Nichols & Warnow, 2008: 761)
Estimated phylogenetic trees are not always binary trees (i.e. every internal node splits into two) because there may insufficient information to fully resolve the relationships between the subfamilies. Furthermore, a network model may be used for languages that have come in contact with each other resulting in phenomenons like borrowing, mixing and creolization to show additional edges indicating dual parentage (Fig. 4.).
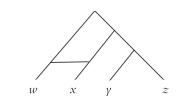
Fig. 4. Tree showing dual parentage
(Nichols & Warnow, 2008: 763)